Immunology is complex. Transciptomics helps. Aganitha DISTILL™ accelerates the journey to insights
“Immunology Is Where Intuition Goes to Die”
– Ed Yong
The mysterious nature of Immunology
Autoimmune diseases challenge the conventional wisdom of immunology. Why does the immune system attack the body it’s meant to protect? Traditional models based on “self” vs “non-self” fall short of explaining these errant responses. Polly Matzinger’s Danger Theory1, proposed in 1994, suggested a shift: immune responses are triggered not by identity, but by signals of damage or danger.
This idea gained fresh support from Waizman et al. (2025)2, who showed that injured skin can prime immune responses at distant sites, a phenomenon they called “remote priming.” When skin was damaged, harmless antigens introduced elsewhere triggered antibody production, but not when the skin was intact. The immune system, it seems, responds to context, not just content. It’s not just what it sees, but how and when it sees it.
From Discovery into Systems-Level Understanding with Single-Cell Transcriptomics
The immune system’s intricate responses, for example, to danger signals from damage or disease, are driven by diverse cell types and complex networks. Traditional methods such as bulk sequencing, flow cytometry suffer from fundamental limitations when studying complex, heterogeneous processes. That is because these bulk approaches average signals across millions of cells, potentially masking rare but critical populations that might mediate critical immune events. Flow cytometry, while providing single-cell resolution, is limited by parameter numbers and relies on predetermined hypotheses about relevant markers. This is where single-cell RNA sequencing (scRNA-seq) becomes indispensable. Single-cell RNA sequencing technology moves beyond average gene changes, capturing the unique expression profiles of individual cells, enabling cell-specific insights, and adding on top of it, pathway enrichment and regulatory networks to create enhanced data-informed insights into autoimmune diseases. This precision allows us to differentiate which specific skin cells release danger signals, which distant immune cells like dendritic cells or rare T cell populations respond, and how these interactions lead to remote priming.
Traditional hypothesis-driven methods, while foundational, often can’t capture the layered, systemic nature of immune responses. To navigate this complexity, immunology now needs a data-first mindset. High-dimensional datasets — from single-cell transcriptomics to spatial transcriptomics — offer a richer lens to uncover patterns we can’t predict upfront. In a field where context is everything, data-driven discovery may be our best path forward.
The integration of scRNA-seq with existing research represents a fundamental shift from hypothesis-driven to discovery-driven science. Traditional approaches require researchers to formulate specific hypotheses about involved cells, molecules, or pathways, then test these hypotheses. While powerful, this approach is limited by existing knowledge and can miss unexpected connections.
While single-cell transcriptomics has enabled a data-driven approach to understanding immunology, it is also laden with a few bottlenecks, such as
- The inability to consistently infer insights across scRNA-seq studies for the same autoimmune disease limits reproducibility, slows therapeutic discovery, and perpetuates siloed research.
While existing tools help, the lack of disease-aware, integration-ready platforms with curated metadata and shared vocabularies remains the core obstacle. Addressing this requires not just better tools, but greater scientific coordination, data standards, and community curation.
- The lack of standardized cell annotations in single-cell RNA sequencing (scRNA-seq) studies is another significant barrier to extracting coherent, actionable insights across studies in the field of autoimmune disease. Even when studies focus on the same disease, they often diverge in how they name, define, and classify cell states—leading to confusion, redundancy, and poor translatability.
Transforming Single-Cell Transcriptomics with Aganitha’s DISTILLTM platform
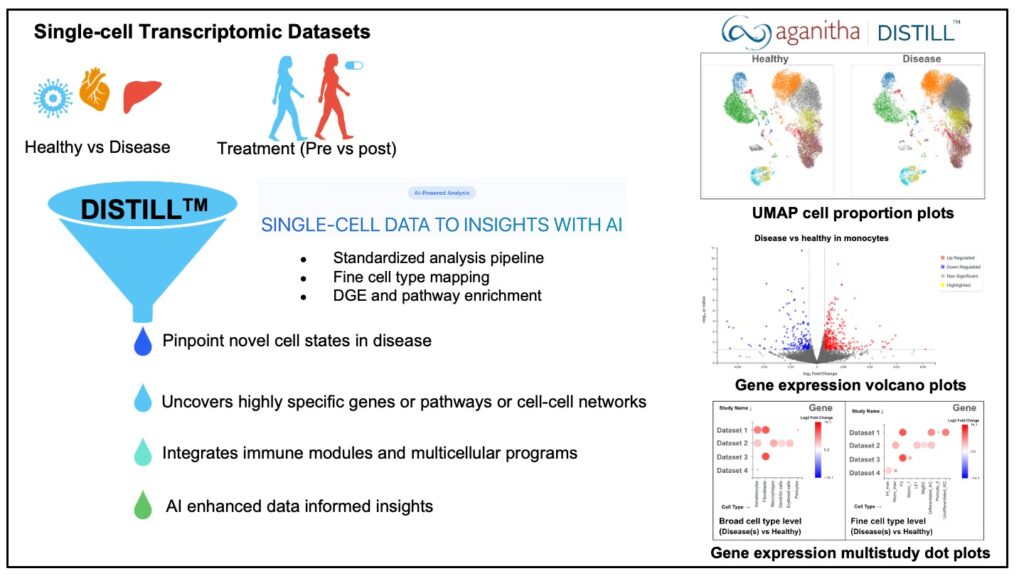
At Aganitha, we make AI work for science. Our single-cell data to insights platform DISTILLTM is designed specifically to make sense of the vast scRNA-seq data for target identification and data-driven MOA insights for autoimmune diseases. This platform can pinpoint novel cell states in inflamed skin, disease, treated or primed tissues, and precisely map how cytokines signal at cellular resolution, and uncovers highly specific genes or pathways, or cell-cell networks that are activated during such states.
In addition, DISTILLTM also integrates immune modules and multicellular programs to obtain precise insights. This cellular precision is crucial for biopharma companies, as it translates observations like the Waizman et al.(2025) “remote priming” phenomenon into actionable, specific drug targets, ultimately accelerating the development of precise therapeutic strategies for autoimmune diseases.
Key insights generated by DISTILLTM include
- Differentially expressed cell proportions and immune module-based analysis
- Differentially expressed gene set analysis and network enrichments
- Cell-cell communication networks between fine-grained cell types
Imagine a scientist eager to understand how CD4+ Th2 cell proportions and their gene expression change in conditions like Atopic Dermatitis, Psoriasis, and other skin disorders. Traditionally, this would involve analyzing each dataset separately, manually extracting insights, and then trying to unify them – a time-consuming process often leading to fragmented research.
With DISTILLTM, this entire process is made easier. The scientist can simultaneously visualize the gene expression profiles and cell type abundance differences across all multiple conditions or datasets, all in a single, integrated view. This not only saves significant time but also fosters a more holistic understanding of immunological changes.
Aganitha’s DISTILL™ platform provides visualization of gene expression across both broad and fine-grained cell types in multiple datasets, offering detailed insight into cellular heterogeneity and its biological significance.
It also produces a multi‐study plot that displays the expression profile of a selected gene across different cell types, disease contexts and datasets, thereby streamlining analysis and promoting a more integrated understanding of immunological variation.
Finally, DISTILL™ also features an integrated LLM capability, transforming complex scRNA-seq data into actionable insights. Simply ask natural language questions like “Which genes are highly expressed in regulatory T cells in psoriatic lesions?” The LLM interprets your query, performs the necessary analysis, and delivers findings in a human-readable format. This intuitive interaction accelerates scientific discovery, empowering more scientists to unlock deeper biological understanding.
Concluding remarks
In a research area as rapidly evolving as immunology, DISTILL™ facilitates the translation of complex single‐cell data into clear, interpretable results. By reducing manual processing steps and integrating diverse datasets, it supports more efficient discovery and a deeper appreciation of immune‐related changes.
DISTILL™ goes beyond this
The scope of DISTILL™ is rapidly expanding. The platform’s evolving capabilities are designed to meet the complex, real-world needs of translational immunology and beyond:
- Multi-omics integration via Igniva™ agents: Connects scRNA-seq data with proteomics, epigenomics, and clinical information to unlock deeper translational insights.
- Cross-functional collaboration: Supports seamless workflows between biologists, clinicians, data scientists, and regulatory teams.
- Iterative, multi-angle analysis: Encourages exploration and re-evaluation across hypotheses, datasets, and disease contexts.
- Meta-studies across boundaries: Enables comparison across species, treatments, and diseases for broader insight.
- LLM-powered insights: Leverages large language models for literature-grounded annotation, natural language querying, and explainable outputs.
We’re just getting started. Stay tuned as we explore more about DISTILL™ in upcoming posts. Contact us to learn more.
- Matzinger P. Tolerance, danger, and the extended family. Annual review of immunology. 1994 Apr 1;12(1):991-1045. ↩︎
- Waizman DA, Brown-Soler I, Martin AL, Ma Y, Zhou K, Israni-Winger K, Zhang C, Medzhitov R, Launay P, Michieletto MF, Henao-Mejia J. Skin damage signals mediate allergic sensitization to spatially unlinked antigen. Science Immunology. 2025 Apr 4;10(106):eadn0688. ↩︎