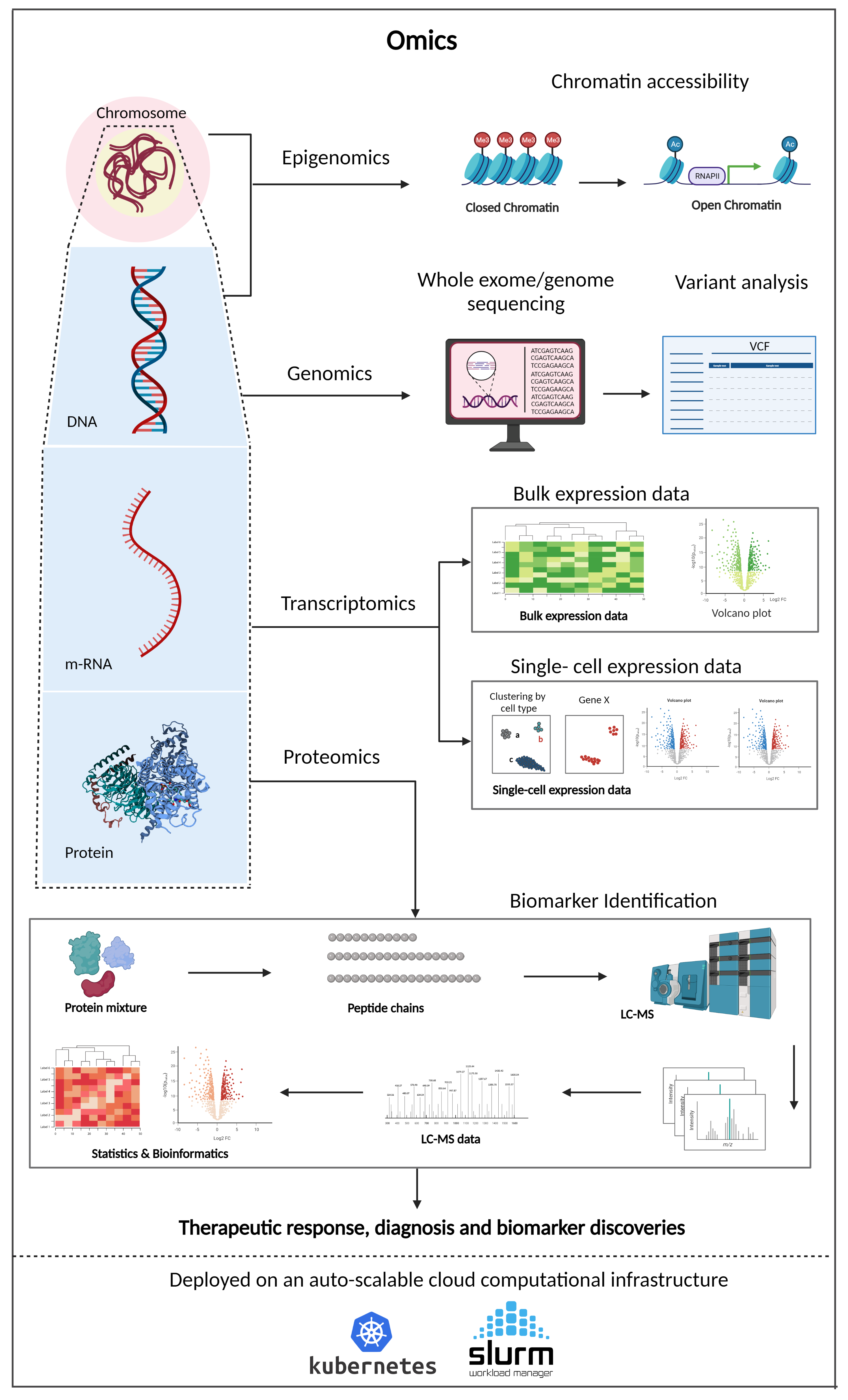
Omics Solutions
R&D Teams in biopharma have a critical dependency on timely availability of high quality insights from data. Satisfying this need has become challenging as multiple trends are converging to produce exponentially larger volumes of multiomics data. For example:
- Several public and private data providers are making datasets available in open and restricted ways
- Sequencing innovations are enabling telomere-to-telomere sequencing
- Human genome research is transitioning from a single reference to a pan genome reference
- Innovations in single cell and spatial transcriptomics producing fine-grained datasets
More pangenomics, spatialomics and other omics data is good provided we can process it all in a reasonable time frame. While several platforms exist to analyze large omics datasets, proprietary platforms are expensive for everyday R&D use, and open source platforms require extensive expertise to be deployed for industrial use.
Our Solution
Omics pipelining on HPC/Cloud as a managed service
An end to end enterprise class, cloud hosted omics pipelining platform with a pre-packaged service offering for quick rollout and managed services for continuous use with the following components:
- Aganitha Omics Kube (AOK) on AWS/GCP/Azure/HPC
- Hail and Cromwell (Open source state of art platforms for omics pipelines from Broad Institute)
- Integrations with Illumina BaseSpace, PacBio SMRT Link, 10x Genomics Cell Ranger, Trans-Proteomic Pipeline from Institute for Systems Biology (Seattle)
- APIs for integration with ELNs such as Benchling
Highlights
Key components & strengths
Proven performance
Cloud & HPC ready
Start quickly with predictable costs
Access to expertise
Extensive domain and technical expertise brought by a cross-functional team to help you focus on science
Outcomes
Reduced costs & time for Omics data analysis added with increased productivity & scalability
Cost and Cycle time reduction
Higher R&D productivity
Unified platform
One-stop solution for Omics analysis, e.g., GWAS, differential gene expression analysis, spatial transcriptomics analysis, protein/metabolite identification and more
Scale quickly
Discover our offerings across the biopharma value chain
Our Solutions
Our Services
Offering services in computational sciences and technology to complement biopharma R&D